The Entrez E-utilities offer a suite of tools that enable researchers to automate the search and retrieval of scientific information from PubMed and other databases maintained by the National Center for Biotechnology Information (NCBI). In Lesson 4 we identified an active NIH funded research project at The Ohio State University and generated a list of PMIDs (PubMed Identifiers) associated with each project. In Lesson 5, we will use this list of PMIDs with the Entrez E-utilities to gather the affiliations of each author listed on the corresponding articles. We will also begin to explore regular expressions, a tool used across programming languages for matching and manipulating string data.
Data skills | concepts¶
Working with APIs
Manipulating text
Regular expressions
Learning objectives¶
Locate API documentation and identify key components required to formulate an API request
Parse an API response and store extracted data.
Utilize regular expressions to search, match, and manipulate text.
This tutorial is designed to support multi-session workshops hosted by The Ohio State University Libraries Research Commons. It assumes you already have a basic understanding of Python, including how to iterate through lists and dictionaries to extract data using a for loop. To learn basic Python concepts visit the Python - Mastering the Basics tutorial.
LESSON 5¶
Science is constantly evolving, with new disciplines emerging from interdisciplinary research, technological innovation, and global collaboration. Analyzing research networks can help researchers identify potential collaborators, track emerging fields, and discover new research directions.
One effective way to explore these networks is by examining author affiliations listed in journal publications.
What is EFetch?¶
EFetch is a utility provided by NCBI’s Entrez system that retrieves detailed records for a list of unique identifiers (like PMIDs) from databases such as PubMed.
Where are author affiliations?¶
In PubMed records, author affiliations are embedded in the XML under:
<Author>
<AffiliationInfo>
<Affiliation>...</Affiliation>
</AffiliationInfo>
</Author>Solution
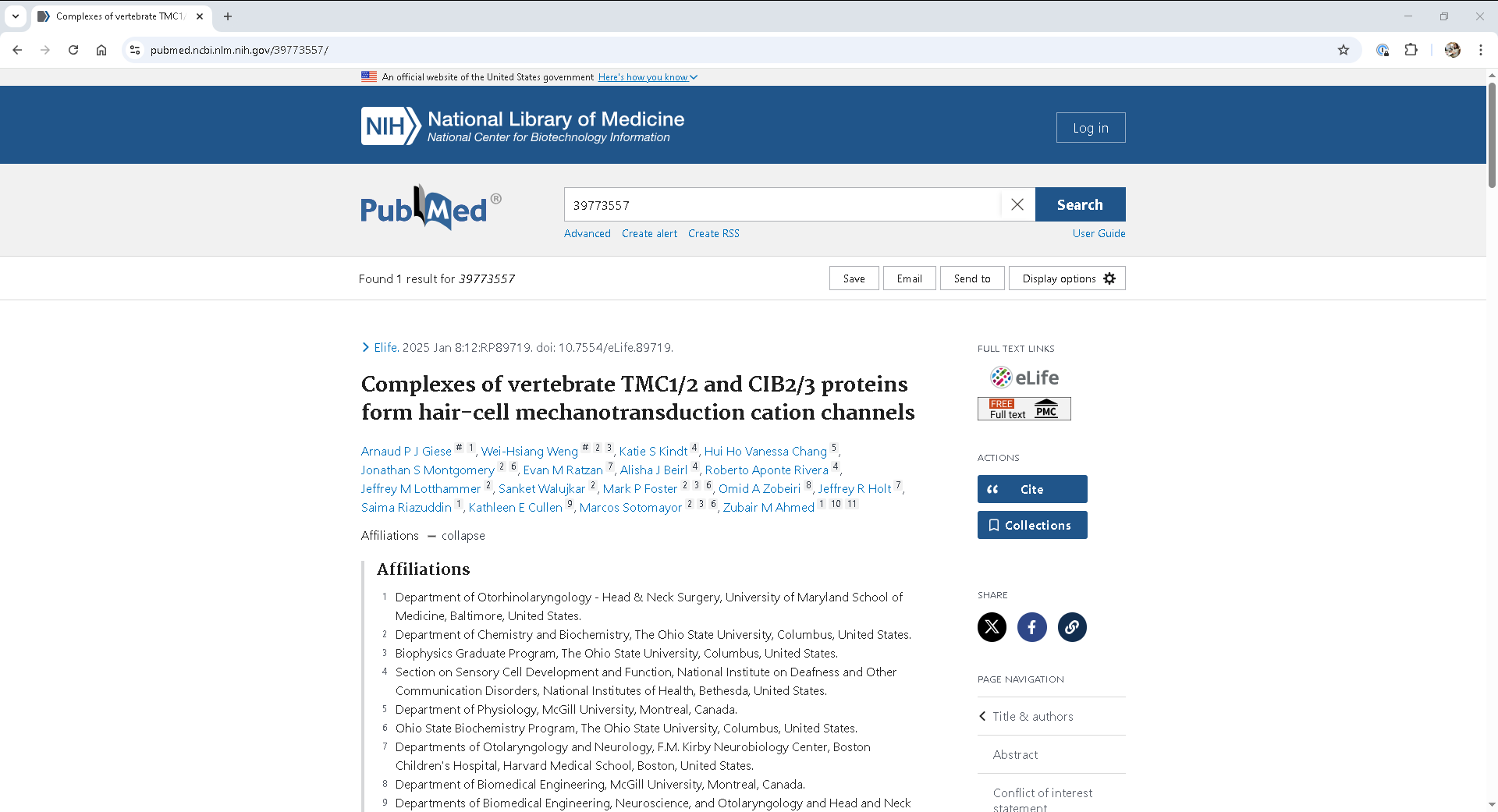
Step 1. Construct an EFetch request¶
To retrieve XML data for a PubMed article, use the following components:
Base URL:
https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgiParameters:
Database name:
?db=pubmedUnique identifier:
&id=39773557API key:
&api_key=INSERT YOUR API KEY HERE
Required parameters for an EFetch request depend on the specific Entrez database you are querying. For PubMed, the default EFetch response format is XML.
To manage request volume, the NCBI enforces rate limits:
Without an API key: 3 requests per second
With an API key: up to 10 requests per second
While you can view a single XML record without an API key, completing the exercises in this tutorial requires one. You can obtain an API key by visiting the Settings page of your NCBI account.
Step 2. Identify Python libraries for project¶
The following Python libraries are needed for this project:
requests– to make HTTP requestspandas– to manage and store dataBeautifulSoup– to parse XML and extract affiliations
Solution
import requests
from bs4 import BeautifulSoup as bs
import pandas as pd
import time
from datetime import date
#1. Create a last_updated variable with today's date.
today = date.today()
last_updated=today
#2. Create list of PMIDs
pmids=['39773557', '39656677', '37398045', '39229161', '39713331', '39315813', '38338688', '36721057', '37322069']
#3.Create a dataframe to store the search results.
author_affiliations=pd.DataFrame(columns=["pmid","name","affiliation","last_updated"])
#4. Use requests, BeautifulSoup, and the EFetch utility to retrieve author affiliations.
# Store results in a DataFrame.
count=0
for each_record in pmids:
# try:
count += 1
print('starting record '+str(count)+': '+str(each_record))
search_url="https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=pubmed&id="+str(each_record)+"&api_key=INSERT YOUR API KEY HERE"
xml_data=requests.get(search_url).text
soup = bs(xml_data, 'xml')
records=soup.find('PubmedArticle')
pmid=records.PMID.text
authors=records.find_all("Author")
for each_author in authors:
if each_author.LastName != None:
lastname=each_author.LastName.text
else:
lastname=''
if each_author.ForeName != None:
forename=each_author.ForeName.text
else:
forename=''
if lastname != '' and forename != '':
name=lastname+', '+forename
else:
name=''
if each_author.Affiliation != None:
affiliation=each_author.Affiliation.text
else:
affiliation=''
print(f"{name}, {affiliation}")
row={
"pmid": pmid,
"name": name,
"affiliation": affiliation,
"last_updated": last_updated
}
author_info=pd.DataFrame(row, index=[0])
author_affiliations = pd.concat([author_info,author_affiliations], axis=0)
time.sleep(0.15)
#5. Export results to csv
author_affiliations.to_csv('data/pubmed_author_affiliations.csv')Regular Expressions (regex)¶
Analyzing author and affiliation data can be messy due to:
Inconsistent naming conventions
Variations in institutional affiliation formats
Ambiguities in author identify.
Regular expressions (regex) match patterns in text. Often described as wildcards on steroids, regular expressions help:
Validate patterns (e.g., ZIP codes: (^\d{5}$)
Extract variations (e.g., “ha?ematology” matches both “hematology” and “haematology”)
Replace text (e.g., re.sub(r’\bOH\b’, ‘Ohio’, text))
Regular expressions are included in several programming langauges and software programs including Python, JavaScript, and Tableau.
Common Regex Patterns¶
| Pattern | Matches |
|---|---|
[A-Z] | Any uppercase letter |
[a-z] | Any lowercase letter |
[0-9]{5} | Exactly 5 digits |
^Ohio | Starts with “Ohio” |
State$ | Ends with “State” |
ha?ematology | “hematology” or “haematology” |
Ohio State|OSU | “Ohio State” or “OSU” |
Metacharacters are special symbols in regular expressions that represent patterns rather than literal characters. To match them as literal characters, you must escape them with a backslash ( \ ).
Common metacharacters and their functions¶
| Symbol | Meaning | Example |
|---|---|---|
[ ] | A set or range of characters | [a-f] matches any lowercase letter from a to f |
\ | Starts a special sequence | \w matches any word character (letter, digit, or underscore) |
. | Any character except newline | d.g matches “dog”, “dig”, “dug”, etc. |
^ | Start of a string | ^Ohio matches any string that starts with “Ohio” |
$ | End of a string | State$ matches any string that ends with “State” |
.* | Zero or more of the preceding character | h*matology matches “hematology”, “haematology”, etc. |
+ | One or more of the preceding character | spe+d matches “sped”, “speed”, etc. |
? | Zero or one of the preceding character | travel?ling matches “traveling”, “travelling”. etc. |
{ } | Exactly a specified number of repetitions | [0-9]{5} matches any 5-digit number |
( ) | Grouping or capturing | The (Ohio) State University extracts “Ohio” |
| | Logical OR | Ohio State|OSU matches “Ohio State” or “OSU” |
LEARNING RESOURCES¶
regex101
regular expressions 101: build, test, and debug regex is an interactive tool that helps you build, test, and debug regular expressions across multiple programming languages. It lets you test your regex against sample text, provides real-time explanations as you type, and includes a searchable reference for regex syntax.
Learning Regular Expressions

Learning Regular Expressions by Ben Forta is available through the Libraries’ O’Reilly Online Learning collection of technical books and videos. Each chapter is structured as a lesson, guiding you through how to match individual characters or sets of characters, use metacharacters, and more—making it a practical resource for mastering regex step by step.
re module¶
To work with regular expressions in Python, start by importing the built-in re module:
import re🔗 See re module documentation.
Commonly used re methods¶
re.match( )¶
re.match(pattern, string, flags=0)Checks for a match only at the beginning of the string.
Returns a match object if found, otherwise None.
Example:
import pandas as pd
import re
addresses=pd.read_csv('data/pubmed_author_affiliations.csv')
addresses=addresses.dropna(subset='affiliation') #drops rows with null affiliation values
for idx, row in addresses.iloc[0:10].iterrows():
affiliation=str(row.affiliation)
print(affiliation)
osu_match=re.match(r"Ohio State University",affiliation)
if osu_match:
print(f" MATCH {osu_match.group()}: {affiliation}")The Ohio State Biochemistry Program, The Ohio State University, Columbus, OH, 43210, USA. fu.978@osu.edu.
The Ohio State Biochemistry Program, The Ohio State University, Columbus, OH, 43210, USA.
Center for Cancer Metabolism, The Ohio State University Comprehensive Cancer Center, Columbus, OH, 43210, USA.
Department of Biological Chemistry and Pharmacology, The Ohio State University, Columbus, OH, 43210, USA.
Department of Biological Chemistry and Pharmacology, The Ohio State University, Columbus, OH, 43210, USA.
The Ohio State Biochemistry Program, The Ohio State University, Columbus, OH, 43210, USA.
Department of Biological Chemistry and Pharmacology, The Ohio State University, Columbus, OH, USA. fu.978@osu.edu.
Department of Biological Chemistry and Pharmacology, The Ohio State University, Columbus, OH, USA.
Department of Physics, Northeastern University, Boston, MA 02115, USA.
Department of Chemistry and Biochemistry, Center for RNA Biology, Ohio State University, Columbus, OH 43210, USA.
🔗 See re.match() documentation.
re.search( )¶
re.search(pattern, string, flags=0)Scans through the string and returns the first match of the pattern.
Returns a match object or None.
Example:
import pandas as pd
import re
addresses=pd.read_csv('data/pubmed_author_affiliations.csv')
addresses=addresses.dropna(subset='affiliation')
for idx, row in addresses.iloc[0:10].iterrows():
affiliation=str(row.affiliation)
# print(affiliation)
osu_search=re.search(r"The Ohio State University",affiliation)
if osu_search:
print(osu_search.group())
else:
print(f"No match: affiliation = {affiliation}")The Ohio State University
The Ohio State University
The Ohio State University
The Ohio State University
The Ohio State University
The Ohio State University
The Ohio State University
The Ohio State University
No match: affiliation = Department of Physics, Northeastern University, Boston, MA 02115, USA.
No match: affiliation = Department of Chemistry and Biochemistry, Center for RNA Biology, Ohio State University, Columbus, OH 43210, USA.
🔗 See re.search() documentation.
re.findall( )¶
re.findall(pattern, string, flags=0)Returns all non-overlapping matches of the pattern in the string as a list.
Example:
# HOW MANY TORTOISES AND TURTLES ARE IN THIS LIST OF ANIMALS?
import pandas as pd
import re
df=pd.read_csv('data/animals_tortoises.csv')
animals=df.common_name.tolist()
animals=','.join(animals)
print('LIST OF ANIMALS')
print(animals)
tortoises_turtles=re.findall(r"tortoise|turtle", animals)
print('ANSWER')
print(f"There are {len(tortoises_turtles)} tortoises and turtles in this list of animals.")
LIST OF ANIMALS
abyssinian-ground-hornbill,addax,african-clawed-frog,african-pancake-tortoise,african-plated-lizard,aldabr-tortoise,allens-swamp-monkey,alligator-lizard,alligator-snapping-turtle,alpaca
ANSWER
There are 3 tortoises and turtles in this list of animals.
🔗 See re.findall() documentation.
re.sub( )¶
re.sub(pattern, repl, string, count=0, flags=0)Replaces all occurrences of the pattern in the string with the replacement text (
repl).You can limit the number of replacements using the count parameter.
Examples:
# FIND TORTOISES AT THE NATIONAL ZOO AND REPLACE THE COMMON_NAME WITH "SLOW TORTOISE"
import pandas as pd
import re
df=pd.read_csv('data/animals_tortoises.csv')
animals=df.common_name.tolist()
animals=','.join(animals)
pattern="tortoise|turtle"
tortoises_slow=re.sub(pattern,"SLOW TORTOISE,",animals)
tortoises_slow'abyssinian-ground-hornbill,addax,african-clawed-frog,african-pancake-SLOW TORTOISE,,african-plated-lizard,aldabr-SLOW TORTOISE,,allens-swamp-monkey,alligator-lizard,alligator-snapping-SLOW TORTOISE,,alpaca'🔗 See re.sub() documentation.
Solution
import requests
from bs4 import BeautifulSoup as bs
import pandas as pd
import time
from datetime import date
import re
#1. Create a last_updated variable with today's date.
today = date.today()
last_updated=today
#2. Create list of PMIDs
pmids=['39773557', '39656677', '37398045', '39229161', '39713331', '39315813', '38338688', '36721057', '37322069']
#3.Create a dataframe to store the search results.
author_affiliations=pd.DataFrame(columns=["pmid","name","affiliation","institution","last_updated"])
#4. Use requests, BeautifulSoup, and the EFetch utility to retrieve author affiliations.
# Store results in a DataFrame.
count=0
for each_record in pmids:
# try:
count += 1
print('starting record '+str(count)+': '+str(each_record))
search_url="https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=pubmed&id="+str(each_record)+"&api_key=INSERT API KEY HERE"
xml_data=requests.get(search_url).text
soup = bs(xml_data, 'xml')
records=soup.find('PubmedArticle')
pmid=records.PMID.text
authors=records.find_all("Author")
for each_author in authors:
if each_author.LastName != None:
lastname=each_author.LastName.text
else:
lastname=''
if each_author.ForeName != None:
forename=each_author.ForeName.text
else:
forename=''
if lastname != '' and forename != '':
name=lastname+', '+forename
else:
name=''
if each_author.Affiliation != None:
affiliation=each_author.Affiliation.text
ohio_state=re.search(r"Ohio State", affiliation)
harvard_medical_school=re.search(r"Harvard Medical School", affiliation)
institut_genetique=re.search(r"Institut de Génétique et de Biologie Moléculaire et Cellulaire", affiliation)
johns_hopkins=re.search(r"Johns Hopkins University", affiliation)
mcgill=re.search(r"McGill University", affiliation)
nci=re.search(r"National Cancer Institute", affiliation)
nidcd=re.search(r"National Institute on Deafness and Other Communication Disorders", affiliation)
northeastern=re.search(r"Northeastern University", affiliation)
u_bristol=re.search(r"University of Bristol", affiliation)
u_maryland=re.search(r"University of Maryland", affiliation)
u_virginia=re.search(r"University of Virginia", affiliation)
vicosa=re.search(r"Universidade Federal de Viçosa", affiliation)
if ohio_state:
institution="The Ohio State University"
elif harvard_medical_school:
institution=harvard_medical_school.group()
elif institut_genetique:
institution=institut_genetique.group()
elif johns_hopkins:
institution=johns_hopkins.group()
elif mcgill:
institution=mcgill.group()
elif nci:
institution=nci.group()
elif nidcd:
institution=nidcd.group()
elif northeastern:
institution=northeastern.group()
elif u_bristol:
institution=u_bristol.group()
elif u_maryland:
institution=u_maryland.group()
elif u_virginia:
institution=u_virginia.group()
elif vicosa:
institution=vicosa.group()
else:
affiliation=''
print(f"{name}, {affiliation}")
row={
"pmid": pmid,
"name": name,
"affiliation": affiliation,
"institution": institution,
"last_updated": last_updated
}
author_info=pd.DataFrame(row, index=[0])
author_affiliations = pd.concat([author_info,author_affiliations], axis=0)
time.sleep(0.15)
#5. Export results to csv
author_affiliations.to_csv('pubmed_author_affiliations.csv')